Introduction
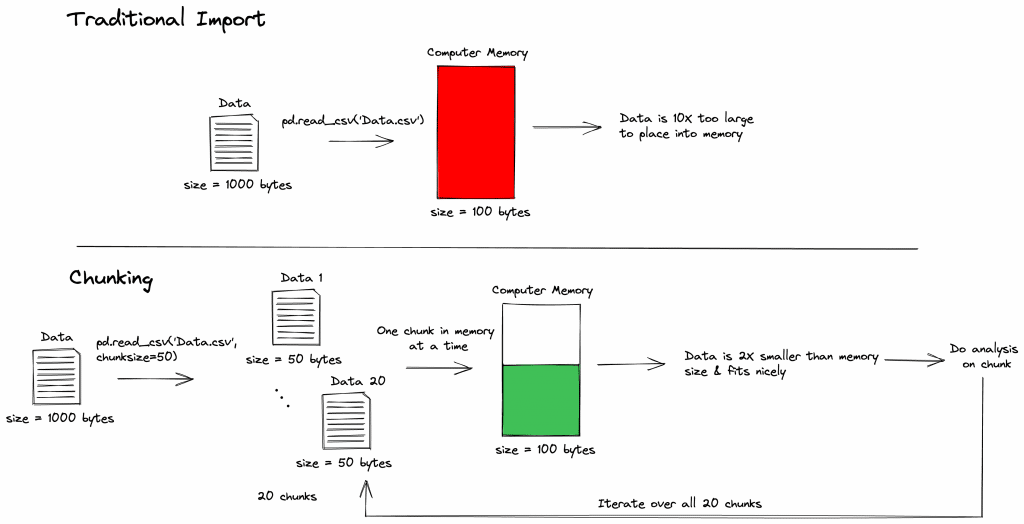
In data science you’ll be confronted with many challenging problems to solve. One of the main challenge areas you encounter is the data preparation stage. So much of effective data science is learning how to work efficiently with data. One of the very first steps of working with data is loading the data. Sounds simple right? Think again. In many situations datasets are too large to fit onto your computer’s local memory. So, what do you do if the dataset you’re working with doesn’t fit into local memory? That is where chunking comes into play.
Chunking is splitting up your large dataset into small datasets. This allows you to perform your analysis pipeline on smaller amounts of data that fit into your computer’s memory. Below you can see a figure that represents the overall idea of chunking and what it solves.
Also Read: Introduction to Long Short Term Memory (LSTM)
In this article, we cover how to perform chunking using the Python library pandas. Pandas, “is an open source, BSD-licensed library providing high-performance, easy-to-use data structures and data analysis tools.” It’s the de facto library for working with data in python, providing many useful, efficient functions for their data structures.
Basic Usage
To read in chunks in pandas one must understand how pandas does file input/output (IO) and the main functions for file IO in pandas. To familiarize yourself with these functions check out the pandas docs: Input/output. Pandas uses the “read_” convention for file input and “to_” for file output. Meaning all file input and output functions will fall under the following syntax:
import pandas as pd file1 = pd.read_csv("myInputFile.csv") ## File Input file1.to_csv("myOutputFile.csv") ## File Output |
For the particular code block above we are reading the file type “csv” or comma-separated values and will be the main focus of the rest of the article. Other file types that can be read into pandas with this convention are listed in Table 1.
| File Type | Input syntax | Output syntax |
| Flat Files (csv, txt, tsv, etc.) | pd.read_csv() | pd.to_csv() |
| Excel Files | pd.read_excel() | pd.to_excel() |
| JSON Files | pd.read_json() | pd.to_json() |
| HTML Files | pd.read_html() | pd.to_html() |
| XML Files | pd.read_xml() | pd.to_xml() |
| HDFStore Files (HDF5) | pd.read_hdf() | pd.to_csv() Use flat file options |
| Feather Files | pd.read_feather() | pd.to_csv() Use flat file options |
| Parquet Files | pd.read_parquet() | pd.to_csv() Use flat file options |
| ORC Objects | pd.read_orc() | pd.to_csv() Use flat file options |
| SAS files | pd.read_sas() | pd.to_sas() |
| SPSS files | pd.read_spss() | pd.to_csv() Use flat file options |
| SQL Tables | pd.read_sql_table(), pd.read_sql_query(), pd.read_sql() | pd.to_sql() |
| Google BigQuery Connections | pd.read_gbg() | pd.to_csv() Use flat file options |
| STATA files | pd.read_stata() | pd.to_stata() |
Now that you understand the basic syntax for loading and saving data using pandas, let’s take a deeper look at read_csv() and how it can be used to perform chunking. The read_csv() function has over 50 possible options to set by the user, but for most use cases you only need to understand a few basic options, mainly, the filepath and the delimiter parameters. These are necessary for properly reading in the file of interest. Filepath is the location of the file of interest. Delimiter defines what character (usually a comma or tab character) signifies a single cell in the file. For reading in chunks pandas provides a “chunksize” parameter that creates an iterable object that reads in n number of rows in chunks. In the code block below you can learn how to use the “chunksize” parameter to load in an amount of data that will fit into your computer’s memory. For the example, we use numpy to generate random gaussian numbers to create csv data files of shape (10000, 10) or 10,000 rows by 10 columns. Then we load the data with the “chunksize” parameter set to 100 & compare it to the traditional method of loading in pandas dataframes to see if they yield the same result of adding the 2 & 3 columns together.
#!/usr/bin/env python # coding: utf-8
import pandas as pd import numpy as np import os for i in range(200):
data = np.random.normal(size=(10000, 10))
data = pd.DataFrame(data)
data.to_csv("./data/test"+str(i+1).zfill(3)+".csv", sep=",")
testfiles = os.listdir("./data/") testfiles = np.sort(testfiles)
###### Chunking pandas approach #######
all_data2 = [] ### List to gather all dataframes of results for i in testfiles:
result = [] ### List to gather all series of add_col1_with_col2
with pd.read_csv("data/"+i, sep=",", chunksize=100, index_col=0) as reader:
for chunk in reader:
add_col1_with_col2 = chunk.iloc[:, 1] + chunk.iloc[:, 2]
result.append(add_col1_with_col2)
result = pd.concat(result)
all_data2.append(result)
result2 = pd.concat(all_data2)
###### Traditional pandas approach #######
all_data = [] for i in testfiles:
data2 = pd.read_csv("data/"+i, index_col=0)
all_data.append(data2)
all_data = pd.concat(all_data) result1 = all_data.iloc[:, 1] + all_data.iloc[:, 2]
### Results are the same np.unique(result1 == result2)
#### OUTPUT #### array([ True])
As we can see from the output from both methods yield the same result, which means that the chunking method works the same, but allowing you to work with the data in a memory efficient manner. Let’s go over what we did with both methods. First, in the chunking methods we use the read_csv() function with the chunksize parameter set to 100 as an iterator call “reader”. The iterator gives us the “get_chunk()” method as chunk. We iterate through the chunks and added the second and third columns. We append the results to a list and make a DataFrame with pd.concat(). Then we iterate through all the files and do the same, get the results of each file and saving them in another list. This list we combine again using pd.concat(). In the second way, we simply open all the files and append them together using pd.concat(). Then we do the math on all the data at once. We can do this because we are using a dataset that fits in memory as an example. Then we check if the chunking method yields the same result as the traditional method and it does. This examples allows us to see the utility of the pandas “chunk_size” option.
Also Read: Coding for Kids: How to Make Pacman on Scratch
Also Consider Dask
When working with large files there have been many innovations that could also help you perform the same tasks in an easier way. One such innovation is the Python library Dask. Dask is a flexible library for parallel computing. It has many great features that can be used on your local machine as well. For instance, you can use dask dataframes in the same way as you use pandas dataframe, but it will always load into memory! How, you ask? This is because dask doesn’t actually load the data into memory until it’s explicitly told! dask uses a “compute()” function that you call when you’re ready to execute some computation. This way you can set up your code in a way that will run quickly and fit in memory. Check out the video from dask that explains the basics of dask dataframes.
Conclusion: Pandas and Large DataFrames
In this article, we demonstrated how and when to use the pandas chunking function to fit large datasets into memory. We compared it to the loading a dataframe without chunk and determined that chunking gives the same result as without, but with the added benefit of fitting within memory. We also covered how you could use dask dataframes as an alternative to the pandas chunking method for more advanced use cases.
Give the utility of working with large datasets, what are your favorite libraries or methods for working with large datasets? Leave a comment below and let us know your thoughts!
